Graph-Based AI for Structural Biology
High-performance ML pipelines, protein structure networks, and agentic AI systems for real scientific work.

High-performance ML pipelines, protein structure networks, and agentic AI systems for real scientific work.

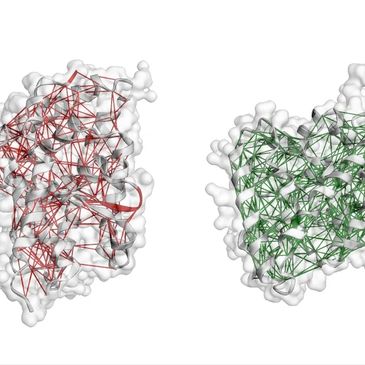
Automated pipelines for constructing and analyzing residue-level interaction graphs, centrality metrics, and allosteric pathways.

GraphSAGE, GAT, and GCN models tailored to protein systems, with interpretability solutions such as Integrated Gradients and saliency.
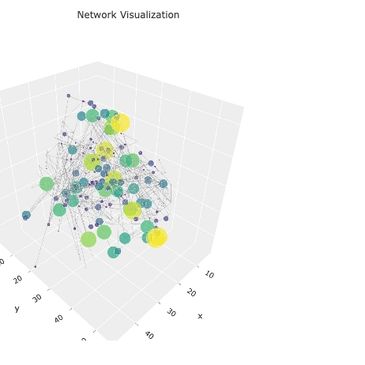
Dash/Flask apps, PyMOL plugins, and visualization workflows for structural data, network topology, and ligand interactions.

Memory-aware agents, computational reasoning tools, and high-performance local ML workflows for scientific discovery.
A PyMOL plugin for generating protein structure networks (PSNs) from PDB or CIF files. The tool computes edge definitions, node metrics, betweenness centrality, and pathway topologies, producing graph outputs ready for downstream ML workflows or GNN-based analysis.
Built and analyzed protein structure networks across multiple LRH-1 states to identify distinct allosteric communication routes associated with ligand efficacy. Network differences between high and low ΔΔG structures revealed Helix 6 as a key regulatory conduit for LRH-1 activation.
Built an 8-head GAT model that learns discriminative patterns in LRH-1 protein structure graphs, classifying conformations associated with high vs. low free-energy states. Integrates saliency and Integrated Gradients to produce interpretable residue influence maps for understanding structural regulation.
A project exploring whether Word2Vec-trained residue sequences ordered by pocket geometry can outperform pooled sequence baselines (ESM, ProtBERT) for kinase binding-site classification. A step toward unifying NLP-style token embeddings with graph-based drug discovery workflows.
A purpose-built machine featuring dual Xeon processors, 256GB ECC RAM, and an RTX 3090, optimized for GNN training, PSN analytics, and memory-aware agentic systems. This workstation supports low-latency inference, heavy graph workloads, and full local autonomy for scientific ML development.
We use cookies to analyze website traffic and optimize your website experience. By accepting our use of cookies, your data will be aggregated with all other user data.